How whale urine benefits the ocean ecosystem
A “great whale conveyor belt”
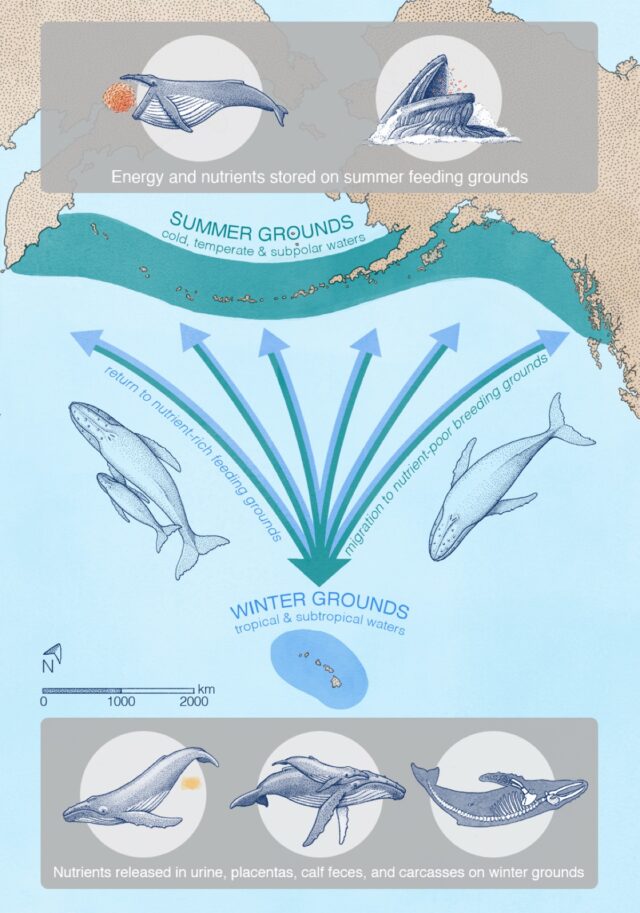
Credit: A. Boersma
Migrating whales typically gorge in summers at higher latitudes to build up energy reserves to make the long migration to lower latitudes. It’s still unclear exactly why the whales migrate, but it’s likely that pregnant females in particular find it more beneficial to give birth and nurse their young in warm, shallow, sheltered areas—perhaps to protect their offspring from predators like killer whales. Warmer waters also keep the whale calves warm as they gradually develop their insulating layers of blubber. Some scientists think that whales might also migrate to molt their skin in those same warm, shallow waters.
Roman et al. examined publicly available spatial data for whale feeding and breeding grounds, augmented with sightings from airplane and ship surveys to fill in gaps in the data, then fed that data into their models for calculating nutrient transport. They focused on six species known to migrate seasonally over long distances from higher latitudes to lower latitudes: blue whales, fin whales, gray whales, humpback whales, and North Atlantic and southern right whales.
They found that whales can transport some 4,000 tons of nitrogen each year during their migrations, along with 45,000 tons of biomass—and those numbers could have been three times larger in earlier eras before industrial whaling depleted populations. “We call it the ‘great whale conveyor belt,’” Roman said. “It can also be thought of as a funnel, because whales feed over large areas, but they need to be in a relatively confined space to find a mate, breed, and give birth. At first, the calves don’t have the energy to travel long distances like the moms can.” The study did not include any effects from whales releasing feces or sloughing their skin, which would also contribute to the overall nutrient flux.
“Because of their size, whales are able to do things that no other animal does. They’re living life on a different scale,” said co-author Andrew Pershing, an oceanographer at the nonprofit organization Climate Central. “Nutrients are coming in from outside—and not from a river, but by these migrating animals. It’s super-cool, and changes how we think about ecosystems in the ocean. We don’t think of animals other than humans having an impact on a planetary scale, but the whales really do.”
Nature Communications, 2025. DOI: 10.1038/s41467-025-56123-2 (About DOIs).
How whale urine benefits the ocean ecosystem Read More »