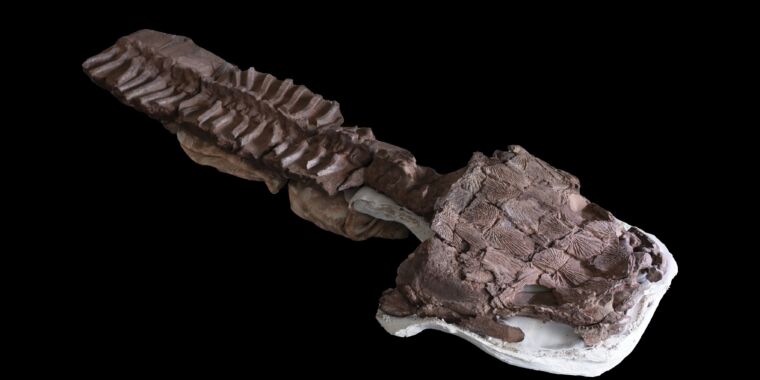
Much of Neanderthal genetic diversity came from modern humans

The basic outline of the interactions between modern humans and Neanderthals is now well established. The two came in contact as modern humans began their major expansion out of Africa, which occurred roughly 60,000 years ago. Humans picked up some Neanderthal DNA through interbreeding, while the Neanderthal population, always fairly small, was swept away by the waves of new arrivals.
But there are some aspects of this big-picture view that don’t entirely line up with the data. While it nicely explains the fact that Neanderthal sequences are far more common in non-African populations, it doesn’t account for the fact that every African population we’ve looked at has some DNA that matches up with Neanderthal DNA.
A study published on Thursday argues that much of this match came about because an early modern human population also left Africa and interbred with Neanderthals. But in this case, the result was to introduce modern human DNA to the Neanderthal population. The study shows that this DNA accounts for a lot of Neanderthals’ genetic diversity, suggesting that their population was even smaller than earlier estimates had suggested.
Out of Africa early
This study isn’t the first to suggest that modern humans and their genes met Neanderthals well in advance of our major out-of-Africa expansion. The key to understanding this is the genome of a Neanderthal from the Altai region of Siberia, which dates from roughly 120,000 years ago. That’s well before modern humans expanded out of Africa, yet its genome has some regions that have excellent matches to the human genome but are absent from the Denisovan lineage.
One explanation for this is that these are segments of Neanderthal DNA that were later picked up by the population that expanded out of Africa. The problem with that view is that most of these sequences also show up in African populations. So, researchers advanced the idea that an ancestral population of modern humans left Africa about 200,000 years ago, and some of its DNA was retained by Siberian Neanderthals. That’s consistent with some fossil finds that place anatomically modern humans in the Mideast at roughly the same time.
There is, however, an alternative explanation: Some of the population that expanded out of Africa 60,000 years ago and picked up Neanderthal DNA migrated back to Africa, taking the Neanderthal DNA with them. That has led to a small bit of the Neanderthal DNA persisting within African populations.
To sort this all out, a research team based at Princeton University focused on the Neanderthal DNA found in Africans, taking advantage of the fact that we now have a much larger array of completed human genomes (approximately 2,000 of them).
The work was based on a simple hypothesis. All of our work on Neanderthal DNA indicates that their population was relatively small, and thus had less genetic diversity than modern humans did. If that’s the case, then the addition of modern human DNA to the Neanderthal population should have boosted its genetic diversity. If so, then the stretches of “Neanderthal” DNA found in African populations should include some of the more diverse regions of the Neanderthal genome.
Much of Neanderthal genetic diversity came from modern humans Read More »